This is documentation for Orange 2.7. For the latest documentation, see Orange 3.
Naive Bayes classifier (bayes)¶
A Naive Bayes classifier is a probabilistic classifier that estimates conditional probabilities of the dependant variable from training data and uses them for classification of new data instances. The algorithm is very fast for discrete features, but runs slower for continuous features.
The following example demonstrates a straightforward invocation of this algorithm:
import Orange
titanic = Orange.data.Table("titanic.tab")
learner = Orange.classification.bayes.NaiveLearner()
classifier = learner(titanic)
for inst in titanic[:5]:
print inst.getclass(), classifier(inst)
- class Orange.classification.bayes.NaiveLearner(adjust_threshold=False, m=0, estimator_constructor=None, conditional_estimator_constructor=None, conditional_estimator_constructor_continuous=None, **argkw)¶
Bases: Orange.classification.Learner
Probabilistic classifier based on applying Bayes’ theorem (from Bayesian statistics) with strong (naive) independence assumptions. Constructor parameters set the corresponding attributes.
- adjust_threshold¶
If set and the class is binary, the classifier’s threshold will be set as to optimize the classification accuracy. The threshold is tuned by observing the probabilities predicted on learning data. Setting it to True can increase the accuracy considerably
- m¶
m for m-estimate. If set, m-estimation of probabilities will be used using M. This attribute is ignored if you also set estimator_constructor.
- estimator_constructor¶
Probability estimator constructor for prior class probabilities. Defaults to RelativeFrequency. Setting this attribute disables the above described attribute m.
- conditional_estimator_constructor¶
Probability estimator constructor for conditional probabilities for discrete features. If omitted, the estimator for prior probabilities will be used.
- conditional_estimator_constructor_continuous¶
Probability estimator constructor for conditional probabilities for continuous features. Defaults to Loess.
- __call__(data, weight=0)¶
Learn from the given table of data instances.
Parameters: Return type:
- class Orange.classification.bayes.NaiveClassifier(base_classifier=None)¶
Bases: Orange.classification.Classifier
Predictor based on calculated probabilities.
- distribution¶
Stores probabilities of classes, i.e. p(C) for each class C.
- estimator¶
An object that returns a probability of class p(C) for a given class C.
- conditional_distributions¶
A list of conditional probabilities.
- conditional_estimators¶
A list of estimators for conditional probabilities.
- adjust_threshold¶
For binary classes, this tells the learner to determine the optimal threshold probability according to 0-1 loss on the training set. For multiple class problems, it has no effect.
- __call__(instance, result_type=0, *args, **kwdargs)¶
Classify a new instance.
Parameters: - instance (Instance) – instance to be classified.
- result_type – GetValue or GetProbabilities or GetBoth
Return type: Value, Distribution or a tuple with both
- __str__()¶
Return classifier in human friendly format.
Examples¶
NaiveLearner can estimate probabilities using relative frequencies or m-estimate:
import Orange
lenses = Orange.data.Table("lenses.tab")
bayes_L = Orange.classification.bayes.NaiveLearner(name="Naive Bayes")
bayesWithM_L = Orange.classification.bayes.NaiveLearner(m=2, name="Naive Bayes w/ m-estimate")
bayes = bayes_L(lenses)
bayesWithM = bayesWithM_L(lenses)
print bayes.conditional_distributions
# prints: <<'pre-presbyopic': <0.625, 0.125, 0.250>, 'presbyopic': <0.750, 0.125, 0.125>, ...>>
print bayesWithM.conditional_distributions
# prints: <<'pre-presbyopic': <0.625, 0.133, 0.242>, 'presbyopic': <0.725, 0.133, 0.142>, ...>>
print bayes.distribution
# prints: <0.625, 0.167, 0.208>
print bayesWithM.distribution
# prints: <0.625, 0.167, 0.208>
Conditional probabilities in an m-estimate based classifier show a shift towards the second class - as compared to probabilities above, where relative frequencies were used. The change in error estimation did not have any effect on apriori probabilities:
import Orange
from Orange.classification import bayes
from Orange.evaluation import testing, scoring
adult = Orange.data.Table("adult_sample.tab")
nb = bayes.NaiveLearner(name="Naive Bayes")
adjusted_nb = bayes.NaiveLearner(adjust_threshold=True, name="Adjusted Naive Bayes")
results = testing.cross_validation([nb, adjusted_nb], adult)
print "%.6f, %.6f" % tuple(scoring.CA(results))
Setting adjust_threshold can improve the results. The classification accuracies of 10-fold cross-validation of a normal naive bayesian classifier, and one with an adjusted threshold:
[0.7901746265516516, 0.8280138859667578]
Probability distributions for continuous features are estimated with Loess.
iris = Orange.data.Table("iris.tab")
nb = Orange.classification.bayes.NaiveLearner(iris)
sepal_length, probabilities = zip(*nb.conditional_distributions[0].items())
p_setosa, p_versicolor, p_virginica = zip(*probabilities)
pylab.xlabel("sepal length")
pylab.ylabel("probability")
pylab.plot(sepal_length, p_setosa, label="setosa", linewidth=2)
pylab.plot(sepal_length, p_versicolor, label="versicolor", linewidth=2)
pylab.plot(sepal_length, p_virginica, label="virginica", linewidth=2)
pylab.legend(loc="best")
pylab.savefig("bayes-iris.png")
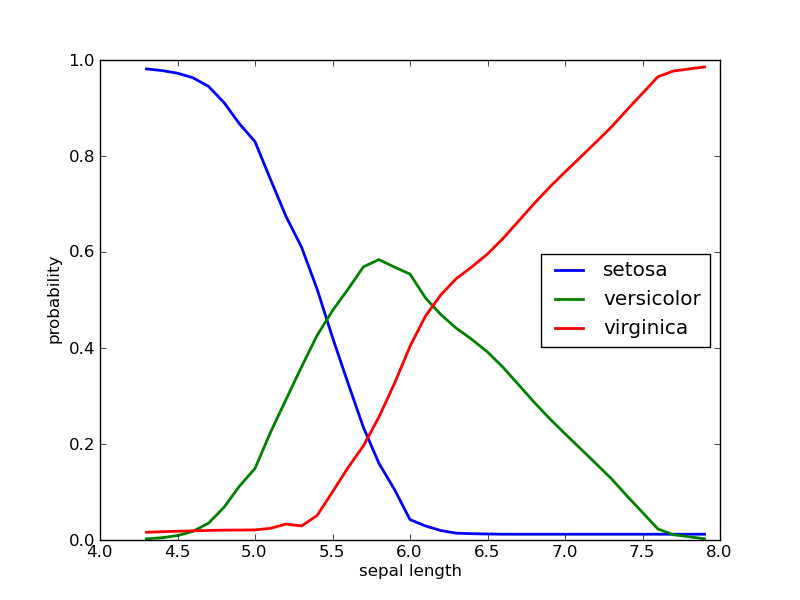
If petal lengths are shorter, the most probable class is “setosa”. Irises with middle petal lengths belong to “versicolor”, while longer petal lengths indicate for “virginica”. Critical values where the decision would change are at about 5.4 and 6.3.
